EASY
Earn 100
How many linear DNA fragments will be produced when a circular plasmid is digested with a restriction enzyme having 3 sites?
(a)4
(b)5
(c)3
(d)2
50% studentsanswered this correctly
Important Questions on Biotechnology - Principles and Processes
MEDIUM
Identify the wrong statement with regard to restriction enzymes.
EASY
Which one of the following represents a palindromic sequence in DNA?
EASY
Identify the palindromic sequence.
MEDIUM
In a restriction enzyme digestion experiment, a fragment of kilobases length is used. This fragment harbors three restriction sites. After the restriction digestion of this fragment with EcoRI, fragments corresponding to base pairs are observed in the agarose gel electrophoresis . Predict the positions of the restriction site in the undigested -fragment which will result in the observed bands in .
EASY
Restriction enzyme work by cleaving
EASY
Select the correct statement from the following:
EASY
Following statements describe the characteristics of restriction endonucleases. Identify the incorrect statement.
EASY
Choose the bacterium which is not a source of :
HARD
A scientist has cloned an 8 Kb fragment of a mouse gene into the Eco RI site of a vector of 6 Kb size. The cloned DNA has no other Eco RI site within. Digestions of the cloned DNA is shown below

Which one of the following sets of DNA fragments generated by digestion with both Eco RI and Bam HI as shown in (iii) is from the gene?
HARD
Eco and Rsa restriction endonucleases require 6bp and 4bp sequences respectively for cleavage. In a 10 kb DNA fragment how many probable cleavage sites are present for these enzymes
EASY
Study the following lists:
| List- | List- | ||
| A. | Eco RI | Vector for transgenic plants | |
| B. | Ti plasmid | Restriction enzyme | |
| C. | pBR | Probe | |
| D. | Reverse transcriptase | RNA dependent DNA synthesis | |
| Artificial plasmid |
The correct match is
MEDIUM
Which ONE of the following statements is INCORRECT about restriction endonucleases?
EASY
Match the following lists:
| List | List | ||
| A) | Hind | pBR | |
| B) | Eco RI | Replication of the plasmid | |
| C) | Hind | First restriction endonuclease | |
| D) | rop codes | E. coli Ry |
The correct match is
EASY
Genomic DNA is digested with a restriction enzyme which is a four base-pair cutter. What is the frequency with which it will cut the DNA assuming a random distribution of bases in the genome?
EASY
Identify the palindromic sequence in the following base sequences
MEDIUM
Explain what are the desired characteristics of an ideal cloning vector used in rDNA technology.
MEDIUM
Identify the sequence which can be cut using .
HARD
A circular plasmid of 10,000 base pairs (bp) is digested with two restriction enzymes, A and B, to produce 3000 bp and 2000 bp bands when visualised on an agarose gel. When digested with one enzyme at a time, only one band is visible at 5000 bp. If the first site for enzyme A (A1) is present at the 100th base, the order in which the remaining sites (A2, B1, and B2) are present is:
MEDIUM
Consider the linear double-stranded DNA shown below. The restriction enzyme sites and the lengths demarcated are shown. This DNA is completely digested with both Eco and Bam restriction enzymes. If the product is analyzed by gel electrophoresis, how many distinct bands would be observed?
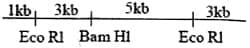
EASY
Which of the following is a restriction endonuclease?

